Menu
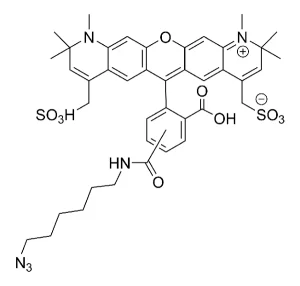
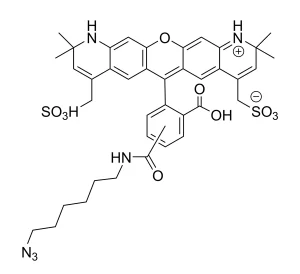
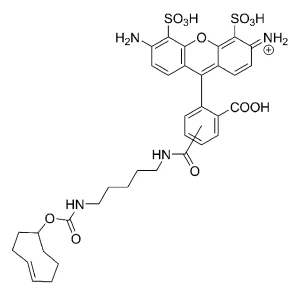
AZDye™ 488 Picolyl Azide is an advanced fluorescent probe that incorporates a copper-chelating motif to raise the effective concentration of Cu(I) at the reaction site to boost the efficiency of the CuAAC reaction, resulting in a faster and more biocompatible CuAAC labeling. Up to 40-fold increase of signal intensity, compared to conventional azides, was reported (see Selected References).
In addition, the use picolyl azides instead of conventional azides allows for at least a tenfold reduction in the concentration of the copper catalyst without sacrificing the efficiency of labeling, significantly improving biocompatibility of CuAAC labeling protocol.
In summary, the introduction of a copper-chelating motif into azide probe leads to a substantial increase in the sensitivity and reduced cell toxicity of CuAAC detection alkyne-tagged biomolecules. This will be of special value for the detection of low abundance targets or living system imaging.
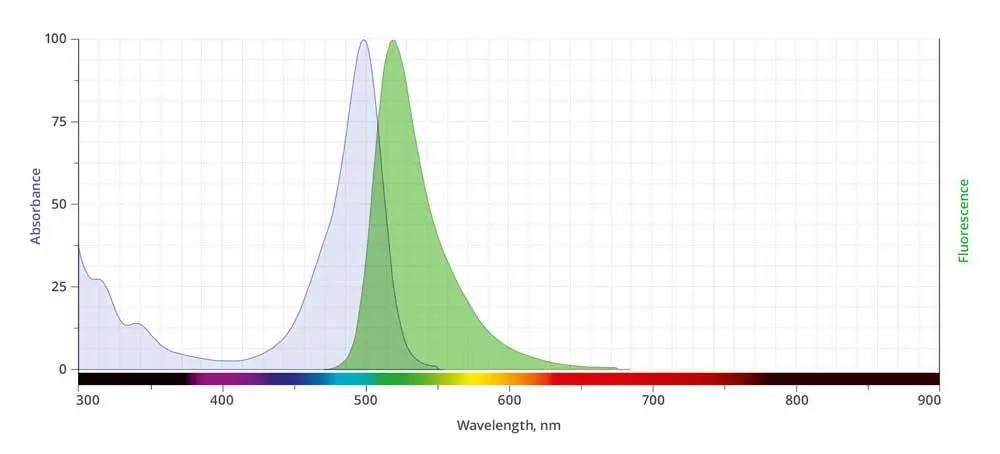
AZDye™ 488 is structurally identical to Alexa Fluor® 488. Its absorption/emission spectra is a perfect match to spectra of many other fluorescent dyes based on sulfonated rhodamine 110 core, including DyLight® 488, Alexa Fluor® 488, and CF® 488A.
| Unit Size | 1 mg, 5 mg, 25 mg |
|---|---|
| Abs/Em Maxima | 494/517 nm |
| Extinction Coefficient | 73.000 |
| Spectrally Similar Dyes | Fluorescein, Alexa Fluor® 488, CF® 488A, DyLight® 488, Atto™ 488 |
| Molecular weight | 736.69 (protonated) |
| CAS | N/A |
| Solubility | Water, DMSO, DMF |
| Appearance | Yellow solid |
| Storage Conditions | -20°C. Desiccate |
| Shipping Conditions | Ambient temperature |
Stay in the Loop
Join Our Online Community
The latest news, tips, and product releases delivered conveniently to your inbox.
We’re social scientists! Let’s connect.
Together we breakthroughTM

©Vector Laboratories, Inc. 2023 All Rights Reserved.
How do I Request a Quote?
To request a quote for products: